Please read the remarks about programming problems for this course.
An adjacency matrix represents the edges in mathematical graph. In this assignment we will consider weighted, undirected, simple graphs. Consider the graph shown in the figure below:
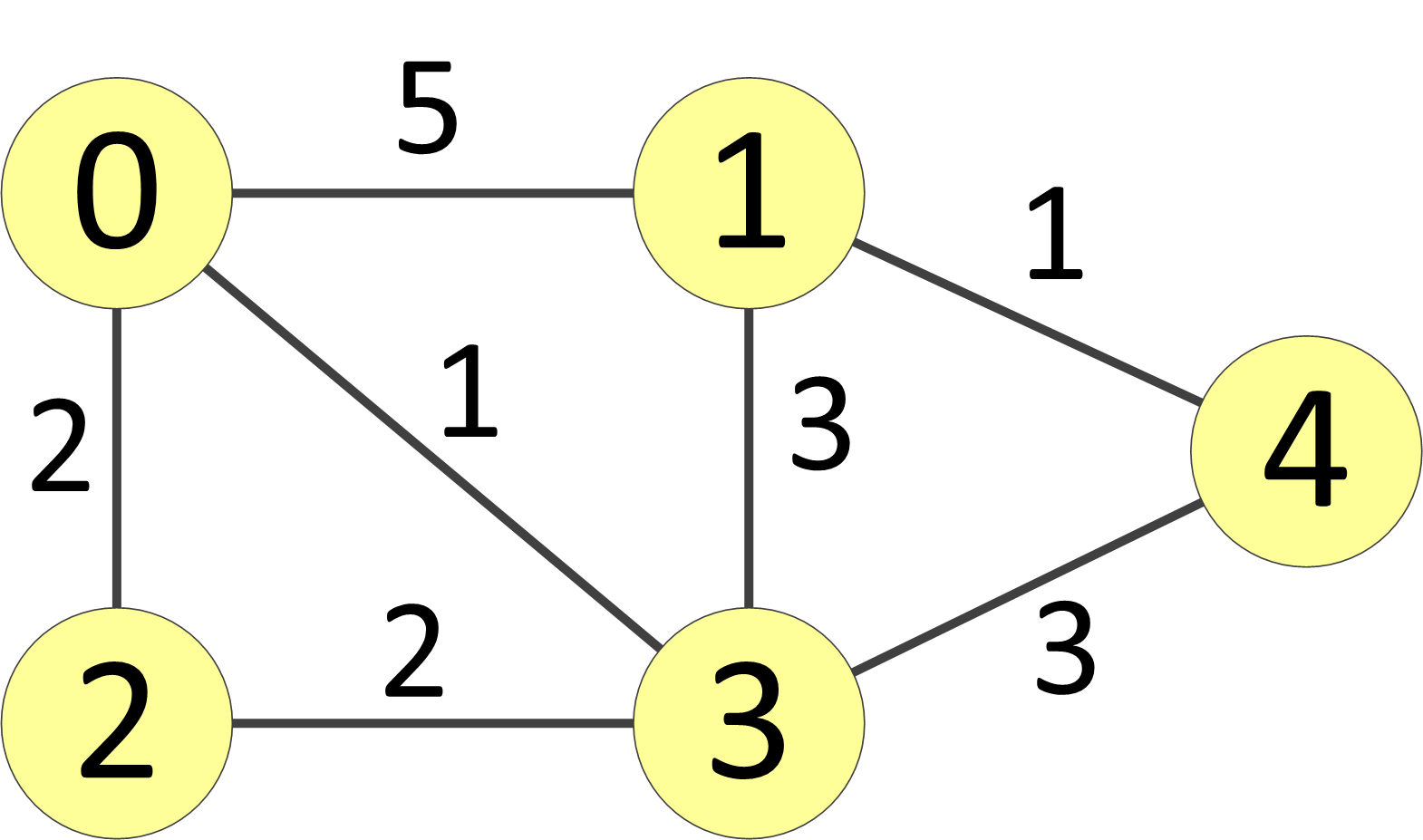
The adjacency matrix for this graph appears here:
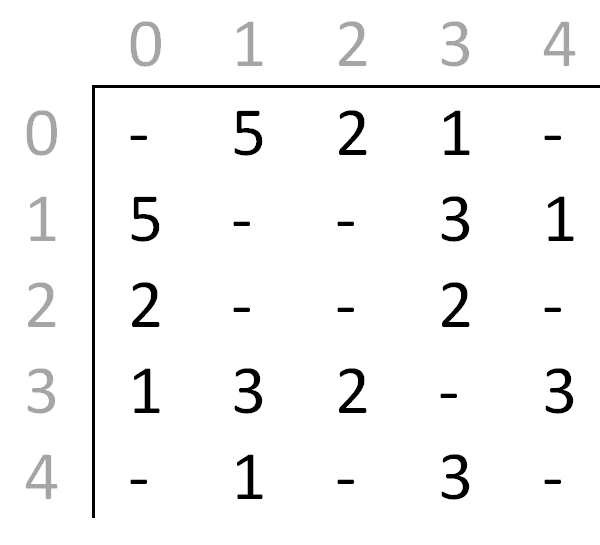
The numbers within the matrix represent edges weights, and the dashes indicate no connection; for example, the element 5 found in row 0, column 1 signifies that an edge of weight 5 directly connects vertex 0 and vertex 1. Note that since the graph is undirected, the matrix is symmetrical about the main diagonal.
Your task is to construct a new matrix which we will call the shortest path martix (SPM) from a given adjacency matrix for an undirected, weighted, simple graph. Rather than containing the weight of the edge connecting two vertices, the entries within a SPM store the weight of a shortest path between two vertices. The following is the SPM for the graph above:

From this SPM we see that a path of length 4 exists between vertices 0 and 1. If this SPM is correct, there can be no other path between these vertices that is shorter.
To simplify and guide your work on this assignment I have provided some support code in the following files:
- graph.h contains the
declarations for a adjacency matrix data structure
(a 2-D
std::vectorof integers), a function that loads data from a text file to create an adjacency matrix, an overloadedoperator<<for conveniently printing an adjacency matrix to an output stream object likestd::cout, and some constants. - graph.cpp implements the functions declared in graph.h.
Your task is to solve the problem of producing the SPM from an adjacency matrix of an undirected, weighted, simple graph. All edge weights will be greater than or equal to zero. You are asked to do so in two different ways:
- We studied Dijkstra's algorithm which computes the shortest path
from a starting
vertex to all other vertices within a graph. One approach to
construct the SPM is to consider each vertex in turn as the
starting vertex and compute the length of the shortest path to all
the other vertices. Because the graph is undirected, when you
compute the shortest path from vertex va to
vertex vb you do not need to compute the
reverse path—simply fill in the corresponding entry in the SPM
reflecting across the main diagonal.
Write a function named
make_spm_dijkstrathat accepts an adjacency matrix and returns the SPM—a new adjacency matrix that holds shortest path lengths instead of edge weights. As described above, the function should apply Dijkstra's algorithm to produce the SPM. To accomplish that, add adijkstra_distancehelper function that accepts an adjacency matrix, a starting vertex, and a destination vertex:- This
dijkstra_distancefunction should return the length of a shortest path between the start vertex and destination vertex. You may use the C++ standardstd::priority_queuedata structure to guide your algorithm. - Since
std::priority_queuedoes not have a method that provides thedecrease_keyoperation, simply enqueue the vertex information with the updated distance estimate back into the priority queue. While this results in multiple copies of information about a particular vertex (only the distance estimates differ), the queue will serve the vertex information with the lowest distance estimate before serving the duplicate original versions. Since you will exit the loop and return once the length of the shortest path to the destination vertex becomes known (you need not compute the shortest path to all other vertices in this case), the extra vertex information entries will not disturb the algorithm's correctness—they simply will occupy a little more space while the algorithm is executing.
- This
- Dijkstra's algorithm is efficient for computing the shortest
path from one starting vertex to any other vertex in a graph;
however, it is not a good fit for computing the SPM.
It works correctly, but we can implement a much faster algorithm.
One problem with using Dijkstra's algorithm to compute the SPM is
this: It recomputes the lengths of segments of the same path multiple
times. In
large graphs this duplicated effort renders the approach
impractical. We
studied a technique that addressed solving problems that involve
multiple overlapping subproblems: dynamic programming.
Write a function named
make_spm_dpthat accepts an adjacency matrix and returns the SPM. The function should use dynamic programming to avoid recomputing portions of paths over and over again. We examined the pseudocode for the dynamic programming algorithm in class; implement it in C++ for this function.
You should implement the functions declared in
spm.h. Place all of your code in a
file named spm.cpp. You may add additional helper
functions (like dijkstra_distance mentioned above) and
accessory data structures to your spm.cpp. Your project
thus should contain the following files:
- graph.h
- graph.cpp
- spm.h
- spm.cpp (you supply this file)
- test_spm.cpp
Important: Your final version of spm.cpp must compile successfully with the unmodified files graph.h and spm.h!
Once your make_spm_dijkstra and
make_spm_dp functions are complete and correct,
time their execution
on graphs containing
15 vertices,
20 vertices,
50 vertices,
100 vertices,
200 vertices,
300 vertices, and
(if you dare) 1,000 vertices.
Report the results of your experiments.
Place all your code to implement the SPM algorithms (including needed support code) within the spm.cpp file. Please submit only your spm.cpp file to eclass.e.southern.edu.